OWL-NETS
What is OWL-NETS?
Our knowledge of the biological mechanisms underlying complex human disease is largely incomplete. While Semantic Web technologies, such as the the Web Ontology Language (OWL), provide powerful techniques for representing existing knowledge, well-established OWL reasoners are unable to account for missing or uncertain knowledge. The application of inductive inference methods, like machine learning and network inference are vital for extending our current knowledge. Therefore, robust methods which facilitate inductive inference on rich OWL-encoded knowledge are needed.
We propose OWL-NETS (NEtwork Transformation for Statistical learning), a novel computational method that reversibly abstracts OWL-encoded biomedical knowledge into a network representation tailored for network inference. Using several examples built with the Open Biomedical Ontologies, we show that OWL-NETS can leverage existing ontology-based knowledge representations and network inference methods to generate novel, biologically-relevant hypotheses. Further, the lossless transformation of OWL-NETS allows for seamless integration of inferred edges back into the original knowledge base, extending its coverage and completeness.
Callahan TJ, Baumgartner Jr WA, Bada M, Stefanski AL, Tripodi I, White EK, Hunter LE. OWL-NETS: Transforming OWL Representations for Improved Network Inference. In Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing 2018 (Vol. 23, p. 133). NIH Public Access. PMC5737627
How Does it Work?
Assumptions
OWL-NETS is implemented in Python (v2.7) and can be run from a simple GUI or from the command line. While primarily developed for use with OWL, the program can be easily extended for use with other Semantic Web technologies by modifying two primary assumptions:
- A knowledge source contains identifiers that directly represent biologically meaningful concepts (e.g., GO:0001525 is the identifier for the biological process of angiogenesis). Within OWL-NETS, biologically meaningful concepts are called “NETS nodes”.
- A knowledge source uses restrictions to specify the existence of biologically important relations between pairs of biological concepts (e.g., proteins restricted to participate in angiogenesis). In OWL, restrictions provide a way to make the definition of a class more specific (e.g., proteins, specifically, protein kinases, participate in phosphorylation). Within OWL-NETS “NETS-edges” represent biologically important relations.
Overview
We describe the OWL-NETS methodology using an example query that investigates disease-associated proteins that participate in angiogenesis (Figure 1). The method takes a SPARQL query as input and outputs a directed OWL-NETS abstraction network, directed OWL representation, or both. To improve processing efficiency, the majority of the computational workload is performed on the input SPARQL query (Steps 1-3) rather than the resulting output.
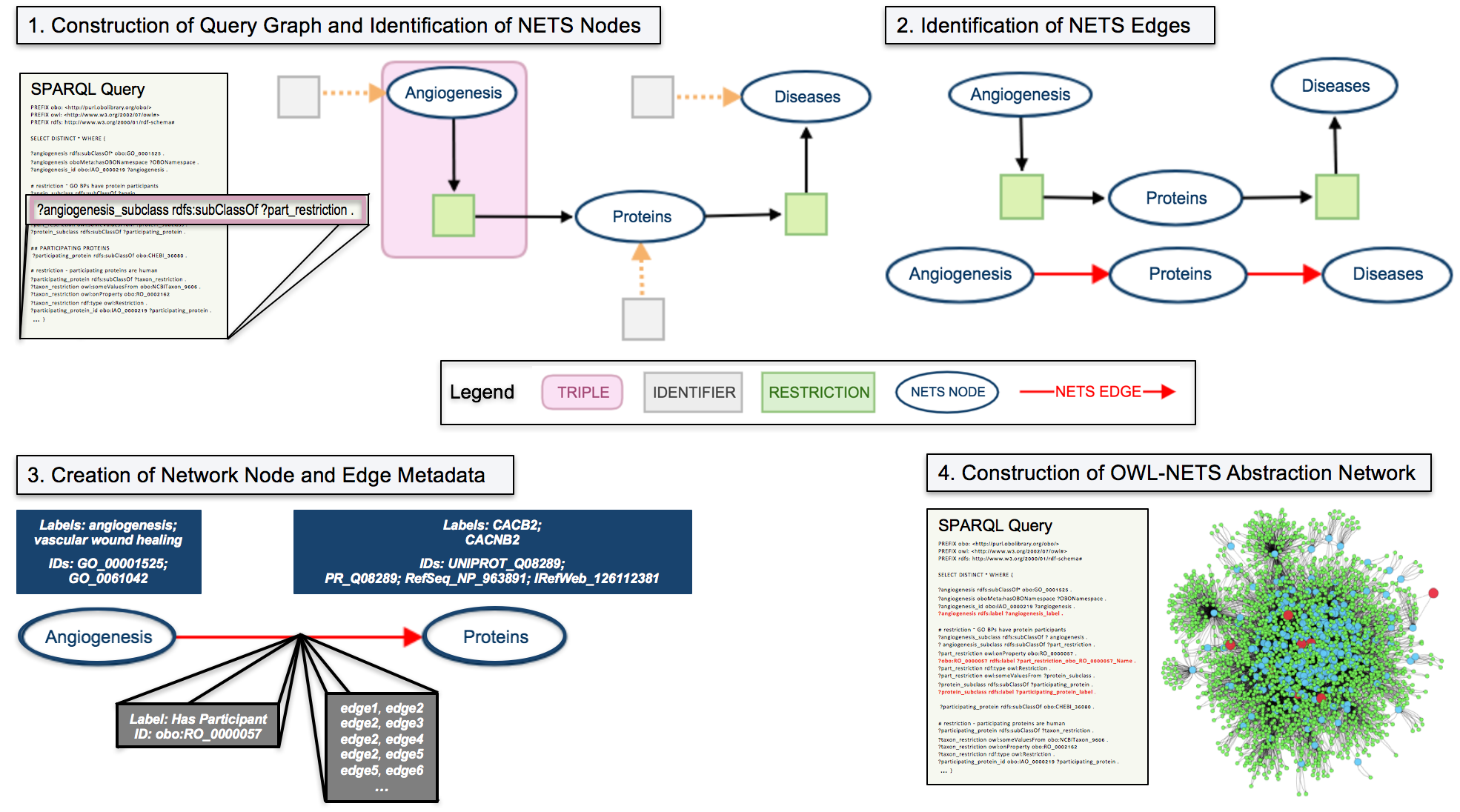
1. Construction of Query Graph and Identification of NETS Nodes: The input SPARQL query is used to create a directed query graph, where each triple (one of which is identified in Figure 1 in the query and in the graph with by pink box) represents a directed edge in the query graph. The graph is then searched for NETS nodes (Figure 1 shows identifier nodes pointing to NETS nodes via dashed orange arrows). NETS nodes in Figure 1 include: Angiogenesis, Proteins, and Diseases.
2. Identification of NETS Edges: The query graph is searched for restrictions (shown in the figure as light green nodes). The NETS node that is reachable from the restriction node’s out-edges is the target of the NETS edge. In Figure 1, one of the green restriction nodes, that is pointed to by Proteins has an out-edge that points towards Diseases, thus an arrow is drawn from Proteins to Diseases. NETS edges are shown in the figure with a red arrow. When both NETS nodes can be reached from a restriction node, arrows pointing in both directions are drawn between the NETS nodes (i.e., nodeA –> nodeB and nodeB –> nodeA).
3. Creation of Network Node and Edge Metadata: Identifiers and labels for each NETS node and edge (shown in the figure as dark blue and gray boxes) are stored as network metadata. This metadata is needed to transform the OWL-NETS abstraction network back into the OWL representation which facilitates the seamless integration of inferred edges back into the knowledge base, extending its coverage and completeness.
4. Construction of OWL-NETS Abstraction Network: Steps 1-3 gather information from the query graph that is needed to construct the OWL-NETS abstraction network. The final step augments the original query with this information. The red lines shown in the example SPARQL query, under Step 4, demonstrate the addition of NETS node and edge metadata to the query. Using this information, the original SPARQL query is updated and run against an endpoint. The endpoint results are then used to construct the OWL-NETS abstraction network.
Biomedical Use Case
| Description | Data Source |
|---|---|
| Human Protein-protein interactions that participate in the biological process of angiogenesis | Gene Ontology; Gene Ontology Annotations |
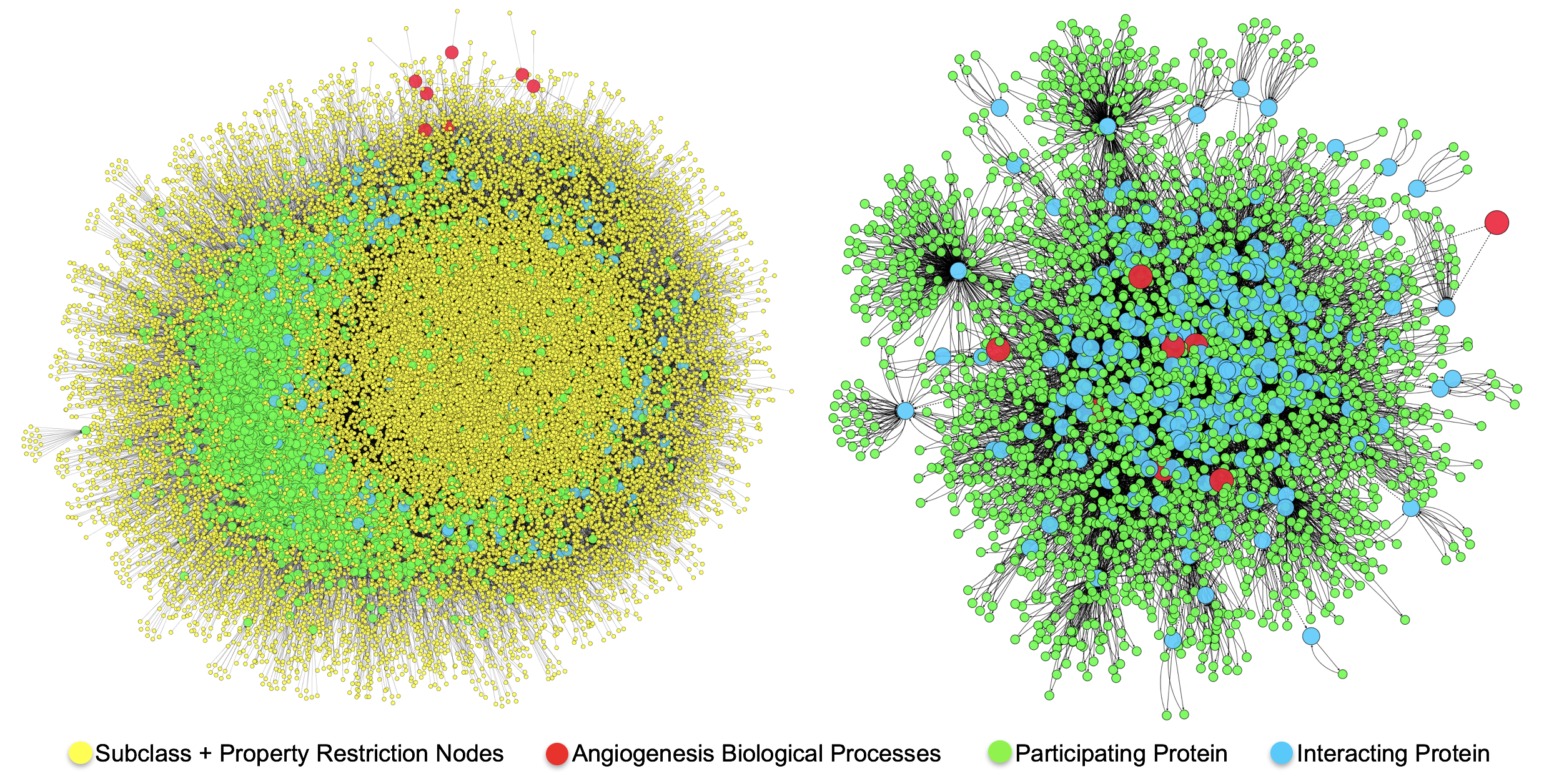 .
.
The image on the left shows the directed OWL network representation of Query 1. The image on the right is the directed absraction network that results when using OWL-NETS. The OWL representation consists of 32,850 nodes and 65,526 edges. The OWL-NETS abstraction network contained 2,2926 nodes and 9,549 edges.